
转录组是连接基因组遗传信息与生物功能的蛋白质组的纽带,转录水平的调控是最重要也是目前研究最广泛的生物体调控方式。转录组测序能够对样品任意时间点或任意条件下的转录组进行测序,拥有精确到单个核苷酸的分辨率。能够动态反映基因转录水平,同时鉴定和定量稀有转录本和正常转录本,并且提供样品特异的转录本序列结构信息。Nanopore三代单分子测序技术为转录组测序提供了新的机遇。
目前转录组测序技术已广泛应用于农学、医学等各个研究领域,包括动植物发育调控、环境适应、免疫互作、基因定位、物种遗传进化及肿瘤与遗传病检测等。Nanopore测序技术在这些方面有无限的潜力。
生物体内复杂多变的转录本是调控基因表达和蛋白质多样性的重要遗传机制,准确鉴定转录本结构,是深入研究基因表达调控模式的基础。Nanopore三代长读长单分子测序技术集二代测序与Pacbio测序的优点于一身,采用该技术,可直接获得全长转录本,准确鉴定基因的可变剪接、APA、融合基因、基因家族等转录本结构信息,同时实现转录本和基因表达定量、差异分析、功能富集分析、差异蛋白互作等。
Oxford Nanopore Technologies (ONT) 是新一代基于纳米孔的单分子实时电信号测序技术,其各平台的测序原理相同。DNA/RNA双链在马达蛋白的带领下与镶嵌在生物膜上的纳米孔蛋白结合并解螺旋,在生物膜两侧电压差的作用下,DNA/RNA链以一定的速率通过纳米孔通道蛋白,由于DNA/RNA链上不同碱基化学性质存在差异,所以当单个碱基或DNA分子通过纳米孔通道时,会引起不同电学信号的变化。通过对这些信号进行检测及对应,即可计算获得相应碱基的类型,完成序列的实时测定。采用Nanopore三代长读长单分子测序技术进行转录组测序,可直接获得全长转录本,准确鉴定基因的可变剪接、APA、融合基因、基因家族和非编码RNA等转录本结构信息,同时实现转录本定量。
部分参考文献:
[1] Deamer D, Akeson M, Branton D. Three decades of nanopore sequencing. Nat Biotechnol, 2016, 34:518-524. https://www.nature.com/articles/nbt.3423
[2] Magi A, Semeraro R, Mingrino A, Giusti B, D’Aurizio R. Nanopore sequencing data analysis: state of the art, applications and challenges. Brief Bioinform, 2017, doi: 10.1093/bib/bbx062. https://www.ncbi.nlm.nih.gov/pubmed/28637243
[3] Jain M, Olsen H E, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol, 2016, 17(1):239. https://www.ncbi.nlm.nih.gov/pubmed/27887629
[4] Bolisetty M T, Rajadinakaran G, Graveley B R. Determining exon connectivity in complex mRNAs by nanopore sequencing. Genome Biology, 2015, 16(1):204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4588896/
[5] Byrne A, Beaudin A E, Olsen H E, et al. Nanoporelong-read RNAseq reveals widespread transcriptional variation among the surfacereceptors of individual B cells[J]. Nat Commun, 2017, 8:16027. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5524981
案例解析
Complete genomic and transcriptional landscape analysis using third-generation sequencing: a case study of Saccharomyces cerevisiae CEN.PK113-7D
研究背景
样品:酵母菌种 CEN.PK113-7D
全长转录组测序:
(1)Nanopore,MinIon,direct RNA,葡萄糖生长条件和乙醇生长条件,各 4 个 生物 学重复(PRJNA398797, SRP116559 )
(2)Illumina,HiSeq 2000,先前研究序列(SRS307298)
分析结果
1.全长转录组数据特征分析
分别对葡萄糖条件和乙醇条件下生长的酵母进行全长转录组测序,葡萄糖生长条件下的酵母 共计获得~509 MB(59X)数据量,包含约 530,000 高质量 reads,其中 N50 为 1150bp;乙醇条 件下的酵母共计获得~623 MB(72X)数据,约 623000 高质量 reads,其中 N50 为 1263 bp。该 技术的比对率为 88%,错误率为 12%,其中超过 70%的转录本为全长转录本,鉴定长度最长转录 本超过 5kb。全长转录本的比例随着转录本长度增加而减小,与转录本的表达量没有明显关系。有22 个转录本是明显高表达的,比如 丰度最高的转录本之一, 的同源 基因,在两个培养条件下表达量都很高;在乙醇条件下特异高表达一些与热休克蛋白、氧化胁迫 相关的基因,其中 编码柠檬酸合酶,说明此时激活了乙醛酸途径;在葡萄糖培养条件下,与 有氧呼吸、核糖体相关的基因表达量较高,符合此条件下酵母生长更快的特征。
2.全长转录组数据差异筛选
通过主成份分析(PCA)发现两组数据有明显的差异,PC1 的贡献率达到 90%。使用 DESeq2做差异分析并对差异基因做 GO 富集分析,结果显示葡萄糖培养条件下的上调基因富集到了与转 录、翻译过程相关的 GO term,与葡萄糖培养条件下生长更快的表型吻合。在乙醇培养条件下, 上调基因主要富集到了 TCA 循环,乙醛酸通路,线粒体电子传递方面。同时,由于营养物质消耗、 毒性代谢物积累,在乙醇条件下很多与胁迫响应,分解代谢,β氧化相关的基因表达量上调。
3.转录组数据结构分析
MinION 的数据中,我们发现相邻的 2 个基因 PTH1 (CENPK0H0281W) 和 ERG9 (CENPK0H0282W) 转录在一个转录本中,有大量的 reads 跨越了基因间区。而 Illumina 的比对结果中, 在两个 ORFs 之间的区域,比对到的 reads 不是完全覆盖的,这种低置信度的信息很可能被忽略。

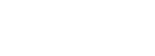

分析平台-150x150.png)
