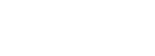蛋白组学是从整体角度分析细胞内动态变化的蛋白质组成、表达水平与修饰状态,了解蛋白质之间的相互作用与联系,揭示蛋白质功能与细胞生命活动规律的新的研究领域。 蛋白质是生命活动的体现者和执行者。要对生命活动有全面深入的认识,必然要在整体、动态、网络的水平对蛋白进行研究。传统的研究方法已经不能满足后基因组时代的需求,高通量,高精度质谱技术的普及使蛋白组学分析成为可能。
蛋白组分析平台可以进行标准分析和个性化分析,其中标准分析包括:蛋白表达分析和蛋白注释分析。设定参数后点击提交进行分析,分析完成后在流程定制页面下生成标准化结题报告,实现一键式生成。
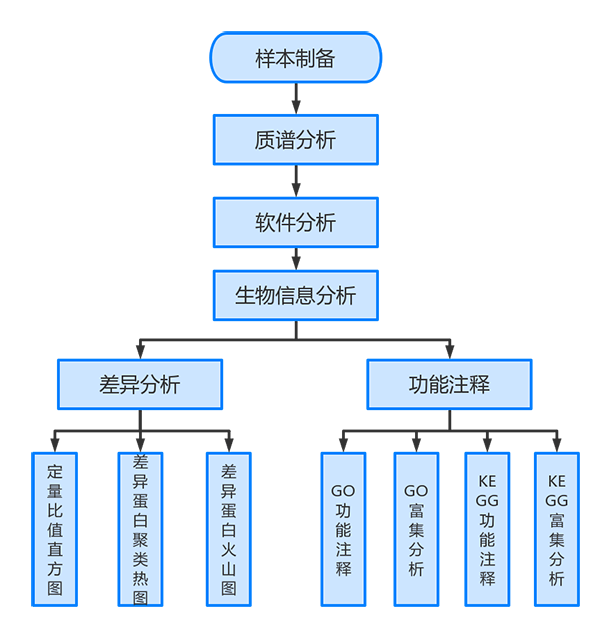
蛋白组分析以定量结果表格作为输入,使用T检验进行差异分析,获取两个分组间的差异表达蛋白。
应用Gene Ontology(简称GO)数据库和KEGG数据库对蛋白进行功能注释,并使用超几何检验,找出与所有蛋白质背景相比,在差异蛋白质中显著富集的GO term和KEGG pathway.
参考文献:
[5] Matrix Science. Introduction to Database Searching using MASCOT. Mascot 2.5.